Diaclone - Editos
Here you will find articles that are related to Diaclone's products and services and its Research & Development activities.
Index
- IL-1ß Monoclonal Antibodies
- TRAIL, the New Grail?
- IL1RAP, a therapeutic target for oncologic treatment
- What you need to know about Exosomes
- Complete Proteome of Covid-19
- Patients’ Immune Battle Against COVID-19
- Covid-19 Latest News
- Anti-CD25
- The Dark Side of CAR-T Therapy: CRS
- Biologically Active Antibodies
- What is the KD of my Antibody?
- Diaclone ELISPOT - It was 30 years ago today...
- Diaclone - The CAM family
- CART poster Diaclone
- Poster IL-8 Phage display
- Diaclone IL-17 Products
IL1 Beta Monoclonal Antibodies
Since 1963, Adler and his colleagues had dedicated their publications to the explanation of the biphasic immune response, this comprised a deep understanding of the early wave of IgM secretion, followed by a later wave of specific IgG occurring in immunized animals.
In fact, they noticed that infection with a T2 bacteriophage of peritoneal exudate cells triggers an immune response in lymph nodes of non-immune animals, when the two populations subsets are exposed together. Although the IgGs were from the allotype of the donor lymph nodes, the IgMs proved to be from the allotype of the donor exudate cells.
Adler et al. therefore questioned the existence of a specific messenger, making the connection between different immune cell subsets.
1. A curious messenger
Since the exudate cells were rich in macrophages, and if the cell of origin was the macrophage that had ingested T2, then the question arose why this cell had to produce messenger RNA relevant for immunoglobulin synthesis when it did not produce such globulins (1).
By the end of 1970, and with the wish to answer this riddle, many teams (2, 3) concluded that the macrophage was involved in antigen processing, antigen concentration, presentation of the antigen to the precursor cell, or in transfer of genetic information specifying immunoglobulin structure to the precursor cell.
A year later, Gery et al. demonstrated that macrophages release one or more mitogenic substances that act on T lymphocytes, greatly potentiating their response to immunogens when activated with lipopolysaccharides. They proposed the term LAF for "lymphocyte-activating factor" for the potentiating activity in question. If they initially recognized that LAF may prove to be heterogeneous, first fractionation studies rapidly suggested that the team was confronted with a single substance (4).
2. The mother of all fevers
At the same moment, Elisha Atkins was working on a different topic. Curious about fever as a consequence of virus and bacterial infection, he was trying to identify and understand the compound involved. This endogenous pyrogen was soon established as a species-specific entity, produced by polymorphonuclear leukocytes and which requires a stimulus to be produced (5). Subjugated by this discovery, Charles Dinarello decided to set-up a radioimmunoassay to detect and accurately measure the circulating leukocytic pyrogen (LP) during fever in humans (6), so replacing the limited Atkins’ bio-assay. Using a macrophage-dependent T-cell assay developed by Lanny Rosenwasser, Charles tested LP on mouse lymphocytes. After two years of repeated testing, he concluded that LP and LAF were the same molecule (7).
3. The very first of its kind
The term interleukin-1 (IL-1) now refers to the originally described endogenous pyrogen and lymphocyte-activating factor. Unlike other cytokine families, the IL-1 family is the mediator of the inflammatory response, at both the receptor and nuclear levels. Members of the IL-1 family of receptors contain activators and suppressors of inflammation and are now the most studied interleukin group.
In 1985, March et al. (8) isolated cDNA libraries from LPS-stimulated macrophages and discovered that IL-1 consists of two distinct proteins, called interleukin-1 alpha (IL-1α) and interleukin-1 beta (IL-1β).
And the latter hadn't said its last word.
4. Let’s listen to the B-side
IL-1β is a cytokine of 269 amino-acids with a molecular weight of 30,7 kDa. It is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis. This cytokine is synthetized as a precursor (proIL-1β) and then cleaved in its biologically active form (17,4 kDa) by Caspase-1, just prior to aspartate residues (9).
Caspase-1 is part of the inflammasome, a complex formed as a result of the recognition of various inflammatory signals by « NOD-like receptor family, pyrin domain containing 3 » (NLRP3) or cryopyrin (10).
Mutations on NLRP3 genes cause spontaneous activation of the NLPR3 inflammasome and lead to excessive IL-1β secretion. This discovery is the basis of a class of chronic inflammatory diseases, uniquely mediated by IL-1β, and known as auto-inflammatory diseases, such as cryopyrin-associated periodic syndrome (CAPS), gout, and DIRA syndrome.
But IL-1β is also part of the complex puzzle of other diseases, playing a role in:
- type 2 diabetes (11)
- cancer (12)
- liver fibrosis (13)
- rheumatoid arthritis (14)
- COVID-19 (15)
and potentially many others….
Diaclone have developed IL-1β monoclonal antibodies (the B-A15 clone, biologically active form) and immunoassays (ELISA and ELISpot kits) to help you monitor the immune responses to diverse infections, diagnose auto-inflammatory diseases, and simply get the best out of your
inflammation research.
Better call Diaclone!
TRAIL, the New Grail?
Did you say TRAIL?
In 1975, Carswell and his team were trying to resolve one of the scientific enigmas of their time: haemorrhagic necrosis. As it may seem odd to put these two terms together, allow me to give you a short explanation.
In the case of haemorrhagic necrosis, tissue damage prevents drainage of venous blood, leading to the haemorrhage. This event interrupts adequate tissue oxygenation, further causing necrosis. A vicious circle then sets in, quickly leading to the patient's death.
Carswell knew that a link had been established between Gram-negative bacteria products, endotoxins (Shear et al., 1953), and human cancer, but curiously enough he observed that endotoxins do not kill tumour cells in culture. He concluded that endotoxins were only indirect factors involved in the evolution of haemorrhagic necrosis conditions. Endotoxins were responsible for the secretion of a substance selectively toxic to tumour cells, the tumour necrosis factor. The TNF superfamily was discovered.
In 1995, Wiley et al. found a new member of the tumour necrosis factor family with the ability to induce fragmentation of Jurkat and U937 cell DNA into soluble multimers. Using Fas ligand as a positive control - which is well known to induce apoptosis - the team concluded then the discovery of a 281 aa TNF-related apoptosis-inducing ligand, and called it TRAIL (1).
But how does TRAIL work?
Like the other members of its family, the TRAIL protein has the characteristics of a type II transmembrane protein arranged in stable homotrimers, presenting no leader sequence. TRAIL has an N-terminal cytoplasmic domain, which is not conserved across family members, while the C-terminal extracellular domain shows significant conservation.
Under its soluble form, TRAIL interacts with five distinct receptors that are encoded by separate genes: four are membrane receptors and one is a soluble receptor called osteoprotegerin (2). However, only type I death receptor TRAIL-R1 (DR4) and TRAIL-R2 (DR5) membrane proteins, which contain an intracellular death domain, can produce apoptotic signals.
The apoptotic signaling pathway of TRAIL is triggered by TRAIL binding to DR4 and DR5, which enables the receptors to homotrimerize, thereby driving formation of the death-inducing signaling complex (DISC) (3). The other TRAIL receptors are named decoy receptors (DcR) and compete with DR4 and DR5 activation, potentially blocking apoptotic signals (4).
Upon ligand stimulation, DR4 and DR5 recruit Fas-associated death domain protein (FADD) through death domain interactions. FADD then recruits pro-caspase-8 and 10, and/or the cellular FLICE (caspase-8)-like inhibitory protein (c-FLIP) to the DISC. c-FLIP competes with caspase-8 for FADD binding in the DISC and inhibits the apoptosis signal (5).
Understanding the first pieces of this complex puzzle, researchers have decided to exploit this finding on TRAIL pathways to develop new cancer treatments.
On the trail of death…to save lives
TRAIL has been rapidly recognized as a promising target for cancer therapy since it can selectively induce apoptosis in tumour cells, but not normal cells. Consequently, different drugs have been elaborated, amongst them:
- Circularly permuted TRAIL (CPT): a recombinant mutant of human Apo2L/TRAIL developed by Beijing Sunbio Biotech Co. Ltd. as a targeted therapy for multiple myeloma and other hematologic malignancies. CPT is a dual pro-apoptotic receptor agonist that directly activates both pro-apoptotic receptors DR4 and DR5.
- Dulanermin (AMG-951): In a similar fashion, Beijing Sunbio Biotech Co. Ltd, Amgen and Genentech (now subsidiary of Roche) developed a rhApo2L/TRAIL with the same mode of action as CPT. Discontinued in 2011.
- Tigatuzumab (CS-1008): a humanized monoclonal antibody targeting DR5, generated by immunization of BALB/c mice with DR5–hIgG1 fusion protein and subsequently humanized by a CDR grafting method, by Daiichi Sankyo. Discontinued in 2013.
- Mapatumumab (HGS-ETR1): a fully human monoclonal antibody targeting DR4, discovered by Cambridge Antibody Technology (now AstraZeneca), and Human Genome Sciences (now GlaxoSmithKline), as a result of the collaboration on CAT's phage display technology.
- Conatumumab (AMG-655): Amgen developed a fully human monoclonal agonist antibody directed against DR5, and licensed it to Takeda. Discontinued in 2011.
- Ganitumab (AMG-479): human monoclonal antibody against type 1 insulin-like growth factor receptor (IGF1R) developed by Amgen. Discontinued in 2012.
- TAS266: agonistic tetravalent single domain Nanobody® targeting the DR5, developed by Ablynx (now Sanofi), licensed to Novartis. Discontinued in 2012.
Despite the involvement of the big names in biotechnology, the thing that may have caught your eye is the large number of drug candidates whose trials have been halted. So, what happened?
The difficulty of TRAIL approach as clinical strategy
In fact, a major problem in clinical trials that use TRAIL-based therapeutics is that cancer cells are either intrinsically resistant or acquire resistance to TRAIL.
After several clinical trials, it appears that:
· importin β1-mediated nuclear localization of DR5 limits the DR5/TRAIL-induced cell death (6)
· low sensitivity to TRAIL also correlated with the expression of anti-apoptotic members of the Bcl-2 family, and overexpression of Bcl-2 can inhibit TRAIL-induced apoptosis (7,8).
· TRAIL resistance has been associated with lipid rafts, where the EGFR pathway is activated, while TRAIL fails to induce effective death-inducing signaling complex formation (9).
It’s therefore possible to develop alternative strategies to modulate the expression of TRAIL, or its death receptors, as novel cancer therapeutics.
This is the case for instance for ONC201 (originally known as TIC10). This selective imipridone small molecule, antagonist of dopamine receptor D2 (DRD2) has been developed by Oncoceutics. Mechanistically, ONC201 induces apoptosis by inactivating AKT and ERK-mediated Foxo3a phosphorylation, resulting in Foxo3a translocation into the nucleus, where Fox3a activates TRAIL transcription (10).
All is not lost, but the understanding of the mechanisms of resistance to TRAIL is still in its infancy and much work needs to be done. Is Oncoceutics the pioneer of a long lineage?
Since 1999, as part of a collaboration with Olivier Micheau's team in Dijon, (11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21) Diaclone has developed its TRAIL portfolio offering cytometry reagents, biologically active antibodies as well as kits for the determination of soluble forms of TRAIL and TRAIL receptors. Diaclone has been an essential support for research in this field for more than 20 years.
Interested to know what Diaclone can do for you in the TRAIL field?
An innovative target in mind? A therapeutic antibody to develop?
Time to contact the team!
IL1RAP, a therapeutic target for oncologic treatment
Introduction
735.
735 autopsy reports.
Report after report, Stephen had a growing conviction that what he was observing could not have happened by chance.
This time, he was sure. A pattern was emerging.
His hypothesis was finally confirmed. His father James had been a successful surgeon, one of the founders of modern scientific pathology, but his discovery could make him even more famous. The impact of what he had in front of him was far greater.
He picked up his stylograph, looked at the date on the calendar, and sat down at his desk. In the light of his candle, he started writing frantically.
“When a plant goes to seed, its seeds are carried in all directions; but they can only live and grow if they fall on congenial soil. While many researchers have been studying ‘the seeds’, the properties of ‘the soils’ may reveal valuable insights into the metastatic peculiarities of cancer cases.” (1)
In 1889, Stephen Paget just laid the first brick of his “soil-seed” theory. He observed that organ distribution of metastases in 735 breast cancers was not random: a tumour cell (“the seed”) needed a proper “soil” to grow.
This “soil” was named “tumor microenvironment”.
But the microenvironment of a tumor is a very complex matter.
1. Identification of a new element of the tumour microenvironment
Within the tumour microenvironment many cellular types are present, intertwined and permanently interacting with each other. Immune cells, blood vessels, collagen, enzymes, glycoproteins and signalling molecules constitute a vast ecosystem.
At the heart of this ecosystem, the cytokine interleukin-1 (IL-1) – via its recognition by type I receptors IL-1R - transmits signal between tumour cell and stromal cell, which stimulates the tumour to grow (2).
To be accurate, it has been found that a type I IL-1R/IL-1RAP complex is required for signalling by all IL-1 agonists and for high affinity binding by IL-1 (3). This subsequent new element, IL1RAP, became the focus of potential novel therapeutic strategies against cancer disease.
But within the large family of cancers (more than 200 different members), Myeloid Leukemia (AML & CML) diseases show specificity.
A specificity that turns out to be an obstacle to therapeutic treatment.
2. The specificity of Myeloid Leukemia and solid tumors
Solid tumours contain a non-homogeneous group of immature cells known as cancer stem cells, which are constantly being formed and which produce tumour-forming mature cancer cells (4). They therefore play a critical role in cancer proliferation, maintenance, progression and recurrence.
In 2010, a team confirmed that IL1RAP was the top candidate for Chronic Myeloid Leukemia (CML) treatment since IL1RAP was a biomarker consistently overexpressed in hematopoietic stem and progenitor cells (HSPC). They were consequently able to generate an IL1RAP-targeting antibody that killed CML CD34+CD38− cells, but not corresponding normal cells, through antibody-dependent cell-mediated cytotoxicity (ADCC). (5)
This concept, which demonstrates a unique concept for the possible eradication of CML stem cells, gave birth to the Swedish biotech company Cantargia.
Only two years later, the overexpression of IL1RAP was demonstrated in Acute Myeloid Leukemia (AML) disease (6) and the same anti-IL1RAP antibody strategy was deployed again.
But if the antibody strategy works, what is the next step?
3. Beyond anti-IL1RAP antibody, anti-IL1RAP-CAR-T treatment
If an anti-IL1RAP antibody can lead to cancer stem cell inhibition and potential destruction, can we simultaneously trigger a T cell-mediated immune response?
Walid Warda, from the team of Christophe Ferrand, at the Blood Transfusion Centre in Besançon, decided to answer this question and go for CAR-T development using an antibody developed by Diaclone (7).
Diaclone first produced and fully characterized a murine monoclonal antibody specific to human IL-1RAP recombinant protein and now offer two different clones B-L43 and B-R58.
The nucleotide sequences encoding the hypervariable regions of this immunoglobulin, constituting the scFv, coupled with those encoding the third generation T activation sequences (CD28-4.1BB-CD3ζ), were cloned into a lentiviral vector.
The obtained construct also includes an inducible cell suicide system consisting of a Rimiducid®-inducible gene encoding caspase-9 (iCASP9 gene), which allows the removal of unwanted activated T-cell CARs (safety switch), and a gene encoding a surface protein (∆CD19) for the identification and selection of T-cell CARs.
Activated T cells were then established from healthy donors or patients and transduced with lentiviral construct. The obtained anti-IL1RAP-CAR-T cells were tested on mice with very good results.
This preclinical work demonstrates for the first time the whole production and validation process of CAR T-cells directed against IL-1RAP-expressing CML stem cells, conducting to tumour regression.
Conclusion
A better understanding of the tumour environment has led to the discovery of the role played by cytokines in cellular communication, in particular interleukin-1 between stromal and tumour cells. One of the components of the IL-1 receptor, IL1-RAP, has thus been identified as a marker of interest in the context of CML treatments, activating NK cells and blocking tumour growth at the same time. Diaclone and Inserm teams have been able to develop a CAR-T therapy to specifically target this biomarker, which has demonstrated great promise during preclinical studies. But the targeting of IL-RAP is still in its infancy, and its role in other diseases - such as Alzheimer's and autoimmune diseases - appears to be prominent.
What you need to know about Exosomes
“Huge opportunities”. “Many hurdles”. “Enormous potential”.
It's just an excerpt of the several terms you'll come across if you search for them on the web.
What am I talking about?
Exosomes. Also referred to as Prostasomes, Tolerosomes, Dexosomes, or Nanovesicles.
Markers of cancer diseases, the basis of broad diagnostic platform, and potential future therapeutics, exosomes have now surpassed 2,000 annual scientific publications.
But why are exosomes so unique?
1) What are exosomes?
Historically, the term "exosomes" comes from the Greek exo "out of" and soma “body”.
In 1981, Trams and his collaborators stumbled upon 40–90-nm exfoliated membrane vesicles issued from cultures of various normal and neoplastic cell lines. Not able to qualify them based upon their physiologic function, they decided to refer them as “exosomes” (1).
The definition of exosomes, as we use it today, was later described by Harding and Johnstone in 1983, following observations in electron microscopy of traffic intracellular transferrin receptor by maturing reticulocytes (2, 3). The authors defined exosomes as microvesicles of endosomal origin, secreted into the extracellular medium after plasma membrane fusion of a multivesiculate endosome.
Up until the 90’s, we thought that exosomes were a means of removing intracellular material. Since then, Grasa Raposo found that B-lymphocytes secrete antigen-presenting exosomes (4) and so a new function was assigned and studied in depth: vector in the intracellular communication. Indeed, the exosomes contain many elements such as lipids, enzymes, proteins and RNAs capable of modifying the physiology of recipient cells.
2) How exosomes are formed?
Contrary to microvesicles, ectosomes, and membrane particles that are formed from the budding of the plasma membrane, the exosomes have an intracellular origin and are released into the extracellular medium after fusion to the plasma membrane of an endosome.
Once endocyted, the surface molecules reach the early endosomes where they are sorted for recycling or degradation. The molecules to be recycled are redirected to the plasma membrane via endosomes. The molecules to be degraded are selectively clustered in areas of the endosomal membrane that invaginate to form intraluminal vesicles (ILVs). These vesicles accumulate in the lumen of the endosomes, which then take the name of multivesicular endosomes (MVEs). MVEs will then merge with the lysosomes. This process leads to the hydrolysis of the vesicles intraluminals and their cargoes.
However, MVEs can also merge with the plasma membrane and thereby release the vesicles they contain. The intraluminal vesicles thus released into the medium extracellular are then called exosomes.
The membrane deformation allowing the formation of ILVs is ensured by the ESCRT machinery (Endosomal Sorting Complex Required for Transport) composed of four multiprotein complexes: ESCRT-0, I, II and III.
3) Why are exosomes so popular?
Exosomes can be found in many biological fluids, including blood, urine (5), saliva (6), breast milk (7), cerebrospinal fluid (8), semen (9), amniotic fluid (10), and ascites (11).
They can be released from a broad spectrum of healthy and tumor cells (12), such as fibroblasts, epithelial cells, neurons, adipocytes, and have even been mentioned in leishmania (13).
And thanks to this diversity of origin, exosomes have been found to play many roles in various biological processes, such as angiogenesis (14, 15, 16, 17), antigen presentation (18, 19, 20), apoptosis (21, 22, 23, 24, 25), and inflammation (26, 27, 28).
Since these processes are key pathways in cancer, neurodegenerative diseases, infections, and autoimmune diseases, exosomes may be the Holy Grail we have been searching for so long.
But searching within the body such tiny elements, which markers can help us?
4) Markers of exosomes
Exosomes carry on their surface characteristic elements:
- Tetraspanins (29), or membrane organizer proteins: CD9 (30), CD37, CD53, CD63, CD81 (30), CD82, and CD151.
- Cell adhesion proteins: integrin, lactadherin, ICAM, EpCAM
- Immunoglobulin supergene family members: MHCI, MHCII, CD86 and CD54
- Lipids: Phosphatidylserine, cholesterol, ceramide and other sphingolipids, LBPA
- Intracellular trafficking elements: RAB, GTPases, annexins
Targeting the membrane proteins on the surface of exosomes is quite useful, as researchers can directly track and isolate compounds of interest with specific monoclonal antibodies linked to magnetic beads or to the affinity column.
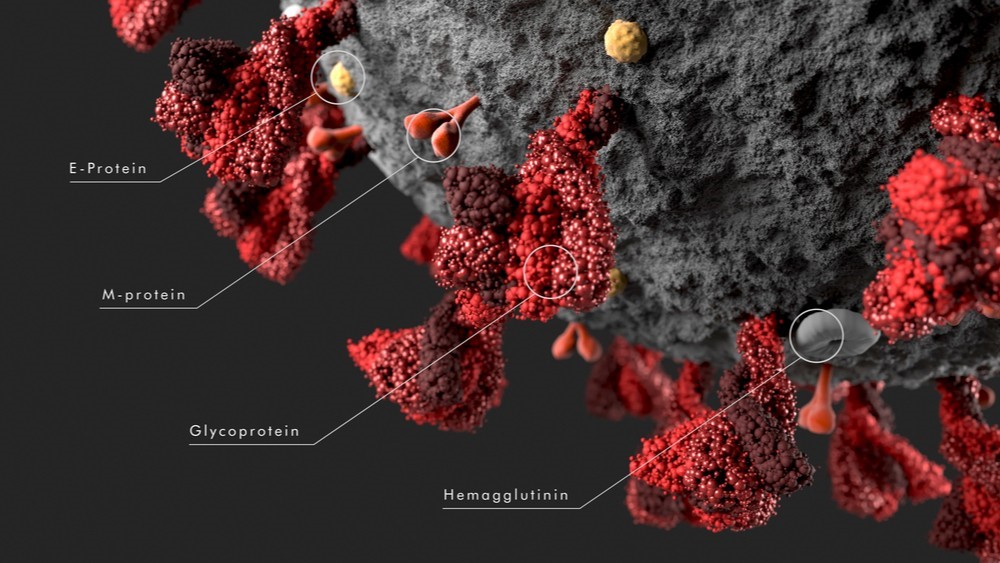
Complete Proteome of COVID-19
The genome of SARS-CoV-2 counts 29,811 nucleotides, encoding for 29 different proteins. The translation of the linear single-stranded RNA conducts to the generation of the following proteome:
- 4 are structural proteins, S, N, M, and E
- 16 proteins are non-structural proteins or NSP: the first 11 are encoded in ORF1a whereas the last 5 are encoded in ORF1b
- 9 are accessory proteins named ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c, and ORF10
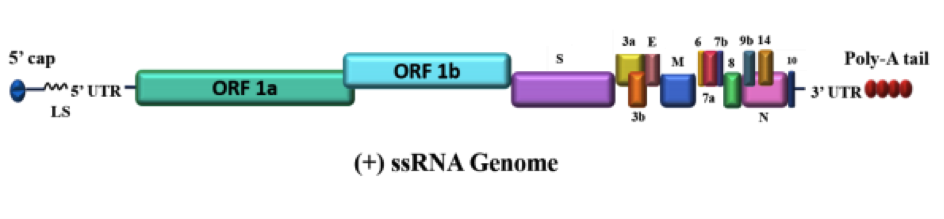
Figure 1. Genome architecture of SARS-CoV-2. SARS-CoV-2 contains a positive single-stranded mRNA as genetic material. The 5’ capped mRNA has a leader sequence (LS), poly-A tail at 3’ end, and 5’ and 3’ UTR. It contains the following genes: ORF1a, ORF1b, Spike (S), ORF3a, ORF3b, Envelope (E), Membrane (M), ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF14, Nucleocapsid (N), and ORF10. Dark Proteome of Newly Emerged SARS-CoV-2 in Comparison with Human and Bat Coronaviruses. Rajanish Giri, Taniya Bhardwaj, Meenakshi Shegane, Bhuvaneshwari R. Gehi, Prateek Kumar, Kundlik Gadhave.
1. Structural proteins of SARS-COV-2
- Spike (S). Trimeric transmembrane spike glycoprotein (1,273 a.a.) precursor is cleaved into S1 and S2 segments in order to bind to ACE2 receptors. A mutation in the SARS-CoV-2 S protein allows the human enzyme called furin to do the proteolytic cleavage into the sequence (PRRARS|V). Since bat CoV doesn’t present this sequence, its apparition may be an explanation for the virus ability to infect humans (1).
- Nucleocapsid (N). Dimer (419 a.a.) that binds viral genome to membrane forming a shell and demonstrates non-specific nucleic acid binding capability (2). Although the overall structure is similar to other reported coronavirus nucleocapsids, the surface electrostatic potential characteristics are unique (3).
- Membrane (M). Matrix protein (222 a.a.) is the most abundant structural component of the virion and defines the shape of the envelope. M protein of SARS-CoV-2 has a triple helix bundle, forms a single 3-transmembrane domain (TM), and is homologous to the prokaryotic sugar transport protein semiSWEET. The advantage and role of sugar transporter-like structures in viruses are still unknown (4). M protein cooperates with Spike during the cell attachment and entry (5).
- Envelope (E). This small membrane protein (75 a.a.) is involved in viral assembly, budding, and pathogenesis. Envelope protein is identical to its counterparts from CoV MP798, CoVZXC21, CoVZC45 and RaTG13. However, a substitution at position 69, and a deletion in position 70 were found (5).
2. Non-structural proteins of SARS-COV-2
Non-Structural Proteins (nsp) are expressed as two long polyproteins (pp1a and pp1ab) then are cleaved into 16 mature smaller proteins by the papain-like protease (PLpro) and the 3-chymotrypsinlike protease (3CLpro, also known as the main protease-Mpro).
- Nsp1 (180 aa): 28 inserts or deletions occurs along its amino-acids sequence, compared to SARS-Cov. The role of nsp1 is believed to be very similar in SARS-Cov and SARS-Cov-2, suppressing type I IFN expression (6).
- Nsp2 (638 aa): nsp 2 interacts with a host protein complex of PHB1 and PHB2 involved in mitochondrial biogenesis (7). Position 321 of this methyltransferase sequence has a polar amino acid while Bat SARS‐like coronavirus has a non-polar amino acid. It can be speculated, that due to this polarity, and potential to form H‐bonds, the glutamine amino acid may confer higher stability to the protein (8).
- Nsp3 (1,945 aa): Nsp3 shuts down host enzymes called PARPs, which prevent viruses from replicating (9). The destabilizing mutation in nsp3 proteins could suggest a potential mechanism differentiating COVID‐2019 from SARS (8).
- Nsp4 (500 aa): Nsp4 critically interacts with nsp3 to rearrange host cell membrane. Only their synergy is able to perform the job (10).
- Nsp5 (306 aa): the main protease of 306 aa cleaves at 11 sites. COVID-19 NSP5 is also highly homologous to SARS NSP5 (96% identity, 98% similarity) (11).
- Nsp6 (290 aa): It interacts with nsp3 and nsp4. Nsp6 presents 7 putative trans-membrane helices like in other coronaviruses. The presence of two mutations affecting the Non-Structural Protein 6 has been recently found (12).
- Nsp7 (83 aa) & Nsp 8 (198 aa): The SARS-coronavirus nsp7+nsp8 primase complex is capable of both de novo initiation and primer extension (13). The complex triggers RNA-synthesizing activity of nsp12 (14). NS7b and NS8, were exclusively conserved among 2019-nCoV, BetaCoV_RaTG, and BatSARS-like Cov. Functional changes in the NS7b and NS8 proteins during evolution may provide important information to explore the human infective property of 2019-nCoV (15).
- Nsp9 (113 aa): Current understanding suggests that Nsp9 is involved in viral genomic RNA reproduction. The structure of the SARS-CoV-2 Nsp9 revealed the high level of structural conservation within the Nsp9 family (16).
- Nsp10 (139 aa): forms a complex with NSP16 to cap viral mRNA transcripts for efficient translation and to evade immune surveillance (17).
- Nsp11 (13 aa): overlapping sequence with nsp10, nsp11 short peptide function is still unknown.
- Nsp12 (932 aa): RNA polymerase complex consisting of the viral RdRp (nsp12) and associated cofactors (nsp7 and nsp8). Alignment of nsp12 for the whole CoV family indicates that the SARS-CoV-2 nsp12 is almost identical to that of the SARS-CoV (96% identity, 98% similarity) (18).
- Nsp13 (601 aa): Like SARS and MERS-Nsp13, the overall structure of SARS-CoV-2 nsp13 adopted a triangular pyramid shape comprising five domains (19). Nsp13 is a helicase that unpacks viral genome material to make it more accessible.
- Nsp14 (527 aa): Nsp14 is the proofreading non-structural protein for normal CoV recombination. Thus, nsp14-ExoN is a key determinant of both high fidelity CoV replication and recombination, and thereby represents a highly-conserved and vulnerable target for virus inhibition and attenuation (20).
- Nsp15 (346 aa): nidoviral RNA uridylate‐specific endoribonuclease (NendoU). While initially Nsp15 was thought to directly participate in viral replication, it was later shown that Nsp15‐deficient coronaviruses were viable and replicating. More recently, it was proposed that NendoU activity of Nsp15 is responsible for the protein interference with the innate immune response, though other studies indicate that the process is independent of the endonuclease activity. There are also suggestions that Nsp15 degrades viral RNA to hide it from the host defences (21).
- Nsp16 (298 aa): forms a complex with NSP10 to cap viral mRNA transcripts for efficient translation and to evade immune surveillance (17).
3. Accessory proteins of SARS-COV-2
- ORF3a (275 aa): the 3a protein is unique to SARS-CoV and is essential for virulence, infectivity, ion channel formation, and virus release (22).
- ORF3b (22 aa): overlapping sequence with 3a, the 3b short peptide is a potent IFN-1 antagonist but allegations have yet to be confirmed. The ORF3b sequences of SARS-CoV-2 are considerably shorter than those of their SARS-CoV orthologs (153.2 ± 0.47 amino acids on average) (23).
- ORF6 (61 aa): IFN-1 antagonist. ORF6 has been shown to be an IFN antagonist that disrupts karyopherin transportation of transcriptions factors like STAT1 (24, 25).
- ORF7a (121 aa): SARS-CoV ORF7a directly binds to BST-2 and inhibits its activity by blocking the glycosylation of BST-2 (26).
- ORF7b (43 aa): overlapping sequence with 7a, ORF7b protein is not only an accessory protein but a structural component of the SARS-CoV virion (27). We haven’t found any reference in SARS-CoV-2 so far.
- ORF8 (121 aa): ORF8 is the most different protein compared to SARS-Cov (30% homology). A 382-nt deletion covering the ORF8 of SARS-CoV-2 has been reported. The deletion also removes the ORF8 transcription-regulatory sequence (TRS), which in turn enhances the downstream transcription of the N gene. (28)
- ORF9b (97 aa): coded within the N gene, ORF9b interacts with a mitochondrial import receptor, Tom70, which acts as an essential adaptor linking MAVS to TBK1/IRF3, resulting in the activation of IRF-3 (29).
- ORF9c (XX aa): coded within the N gene, sigma 2 receptors are hijacked by ORF9c. ORF9c protein was found to interact with multiple proteins that modulate IkB kinase and NF-kB signalling pathway including NLRX1, F2RL1, NDFIP2 (29).
- ORF10 (38 aa): ORF10 does not have any similar proteins in the NCBI repository for SARS-CoV (ORF10 had a premature stop codon in both SARSCoV and BM48-31) and seems unique to SARS-CoV-2 (30).
And you, which one of these proteins are you working on?
Which one would you like to see developed and validated?
You can now contact Diaclone to discuss these topics!
REFERENCES
(1) COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. B. Robson
(2) Biochemical characterization of SARS-CoV-2 nucleocapsid protein Weihong Zeng, Guangfeng Liu, Huan Ma, Dan Zhao, Yunru Yang, Muziying Liu, Ahmed Mohammed, Changcheng Zhao, Yun Yang, Jiajia Xie, Chengchao Ding, Xiaoling Ma, Jianping Weng, Yong Gao, Hongliang He, and Tengchuan Jina
(3) Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B. 1016/j.apsb.2020.04.009.Kang, Sisi & Yang, Mei & Hong, Zhongsi & Zhang, Liping & Huang, Zhaoxia & Chen, Xiaoxue & He, Suhua & Zhou, Ziliang & Zhou, Zhechong & Chen, Qiuyue & Yan, Yan & Zhang, Changsheng & Shan, Hong & Chen, Shoudeng.
(4) Thomas, S. The Structure of the Membrane Protein of SARS-CoV-2 Resembles the Sugar Transporter semiSWEET
(5) Bianchi, Martina & Benvenuto, Domenico & Giovanetti, Marta & Angeletti, Silvia & Ciccozzi, Massimo & Pascarella, Stefano. (2020). Sars-CoV-2 Envelope and Membrane Proteins: Differences from Closely Related Proteins Linked to Cross-species Transmission?
(6) Severe Acute Respiratory Syndrome Coronavirus nsp1 Suppresses Host Gene Expression, Including That of Type I Interferon, in Infected Cells. Krishna Narayanan, Cheng Huang, Kumari Lokugamage, Wataru Kamitani, Tetsuro Ikegami, Chien-Te K. Tseng, Shinji Makino
(7) Comparative Genomic Analysis of Rapidly Evolving SARS CoV-2 Viruses Reveal Mosaic Pattern of Phylogeographical Distribution Roshan Kumar, Helianthous Verma, Nirjara Singhvi , Utkarsh Sood , Vipin Gupta, Mona Singh , Rashmi Kumari, Princy Hira, Shekhar Nagar, Chandni Talwar, Namita Nayyar, Shailly Anand, Charu Dogra Rawat, Mansi Verma, Ram Krishan Negi, Yogendra Singh and Rup Lal.
(8) COVID‐2019: The role of the nsp2 and nsp3 in its pathogenesis. Silvia Angeletti, Domenico Benvenuto, Martina Bianchi, Marta Giovanetti, Stefano Pascarella, Massimo Ciccozzi
(9) https://timesofindia.indiatimes.com/india/a-covid-guide-understanding-the-s-factor/articleshow/75671621.cms
(10) Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Yusuke Sakaiad, Kengo Kawachiab, Yutaka Teradaa, Hiroko Omori, Yoshiharu Matsuur, Wataru Kamitani
(11) Global profiling of SARS-CoV-2 specific IgG/ IgM responses of convalescents using a proteome microarray He-wei Jiang, Yang Li, Hai-nan Zhang, Wei Wang, Dong Men, Xiao Yang, Huan Qi, Jie Zhou, Sheng-ce Tao
(12) Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. Domenico Benvenuto, Silvia Angeletti, Marta Giovanetti, Roberto Cauda, Massimo Ciccozzi, Antonio Cassone.
(13) The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Aartjan J.W. te Velthuis, Sjoerd H. E. van den Worm, and Eric J. Snijder.
(14) One severe acute respiratory syndrome coronavirusprotein complex integrates processive RNApolymerase and exonuclease activitiesLorenzo Subissia, Clara C. Posthumab, Axelle Colleta, Jessika C. Zevenhoven-Dobbeb, Alexander E. Gorbaleny, Etienne Decroly, Eric J. Snijder, Bruno Canarda, and Isabelle Imbert
(15) Nonstructural proteins NS7b and NS8 are likely to be phylogenetically associated with evolution of 2019-nCoV. Fahmi, Muhamad & Kubota, Yukihiko & Ito, Masahiro
(16) Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. D. R. Littler, B. S. Gully, R. N. Colson, J Rossjohn.
(17). Rosas-Lemus, Monica & Minasov, George & Shuvalova, Ludmilla & Inniss, Nicole & Kiryukhina, Olga & Wiersum, Grant & Kim, Youngchang & Jedrzejczak, Robert & Enders, Michael & Jaroszewski, Lukasz & Godzik, Adam & Joachimiak, Andrzej & Satchell, Karla. (2020). The crystal structure of nsp10-nsp16 heterodimer from SARS CoV-2 in complex with S-adenosylmethionine.
(18) Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. AshleighShannona, Nhung Thi-Tuyet Le, Barbara Seliskoa, Cecilia Eydoux, Karine Alvarez, Jean-Claude Guillemot, Etienne Decroly, Olve Peersen, Francois Ferron, BrunoCanard
(19) Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. Muhammad Usman Mirza, Matheus Froeyen.
(20) The coronavirus proofreading exoribonuclease mediates extensive viral recombination. Gribble, Jennifer & Pruijssers, Andrea & Agostini, Maria & Anderson-Daniels, Jordan & Chappell, James & Lu, Xiaotao & Stevens, Laura & Routh, Andrew & Denison, Mark.
(21) Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Youngchang Kim, Robert Jedrzejczak, Natalia I. Maltseva, Mateusz Wilamowski, Michael Endres, Adam Godzik, Karolina Michalska, Andrzej Joachimiak.
(22) SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis - Elio Issa, Georgi Merhi, Balig Panossian, Tamara Salloum, Sima Tokajian
(23) SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is further increased by a naturally occurring elongation variant. Yoriyuki Konno, Izumi Kimura, Keiya Uriu, Masaya Fukushi, Takashi Irie, Yoshio Koyanagi, So Nakagawa, Kei Sato.
(24) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and 278 nucleocapsid proteins function as interferon antagonists. Kopecky-Bromberg, S.A., Martinez-Sobrido, L., Frieman, M., Baric, R.A. & Palese.
(25) Frieman, M., et al. Severe acute respiratory syndrome coronavirus ORF6 antagonizes 280 STAT1 function by sequestering nuclear import factors on the rough endoplasmic 281 reticulum/Golgi membrane. Journal of virology 81, 9812-9824 (2007).
(26) Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J.K. Taylor, C.M. Coleman, S. Postel, J.M. Sisk, J.G. Bernbaum, T. Venkataraman, et al.
(27) The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. Schaecher SR, Mackenzie JM, Pekosz A
(28) Discovery of a 382-nt deletion during the early evolution of SARS-CoV-2. Running title: A SARS-CoV-2 deletion variant. Yvonne CF Su, Danielle E Anderson, Barnaby E Young, Feng Zhu, Martin Linster, Shirin Kalimuddin, Jenny GH Low, Zhuang Yan, Jayanthi Jayakumar, Louisa Sun, Gabriel Z Yan, Ian H Mendenhall, Yee-Sin Leo, David Chien Lye, Lin-Fa Wang, Gavin JD Smith.
(29) A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential DrugRepurposing David E. Gordon, Gwendolyn M. Jang, Mehdi Bouhaddou, Jiewei Xu, Kirsten Obernier, Matthew J. O'Meara, Jeffrey Z. Guo.
(30) On the origin and continuing evolution of SARS-CoV-2. Xiaolu Tang, Changcheng Wu, Xiang Li, Yuhe Song, Xinmin Yao, Xinkai Wu, Yuange Duan, Hong Zhang, Yirong Wang, Zhaohui Qian, Jie Cui, and Jian Lu.
Patients’ Immune Battle Against COVID-19
My stepfather entered the hallway, he seemed troubled by something. He isn’t the kind of man who usually shows his feelings. He used to pass by our house at least twice a week to discuss a while and to barter. I buy fresh eggs from my neighbour and trade them against fresh vegetables my stepfather grows.
But that morning, Luc wasn’t himself. His gaze averted, not the slightest of smile on his lips. He leaned gently towards me and whispered a few words.
His mother had just passed away. It all happened so suddenly. In the space of just a few days.
A brief review of the expected complications
1. Primary battle zone: From fever to lung infection
On the first day it starts with a fever for 94% of us. We feel exhausted in some cases (23%), and face muscle pain like the day after a big workout (15%). And we start coughing after 5 days. A lot, in fact (79% of reported cases) (1).
At this point, severe cases (15%, according to the Chinese CDC) start experiencing polypnea; an increase in the number of breathing cycles per minute and a decrease in the amplitude of breathing movements. In short, you are progressively running out of air.
Doctors are accustomed to polypnea (first described in 1889) and know to search the causes in lung or heart diseases. At the beginning of the pandemic, polypnea was the first symptom observed to attest worsening infection. On January 7th 2020, Chinese scientists released the first X-ray images and sample bronchoalveolar-lavage fluids to finally isolate and sequence viral RNA. This coronavirus was not like the others. Scientists just stumbled upon the newest member of the coronavirus family, the 7th one (2): COVID-19.
And curiously enough, they started observing something else that was occurring in patients.
2. Multiple battlefields: Beyond the lung infection
By mid-January 2020, in the hospital of Wuhan, the dialysis machines began to run out.
In the emergency rooms treating COVID-19, renal distress was raging: 23 patients out of a total of 85 exhibited acute renal failure (3).
In February, another hospital reported heart-related complications: 23% of the patients were suffering arrythmia, and 10% of them faced acute cardiac injury (4).
Lungs. Kidneys. Heart. Where does the list end?
These abnormalities followed on from one another to finally reach the neurological system. Acute cerebrovascular events spread among severely infected patients (36% of a total of 214 patients). Loss of smell, headaches, ataxia and nerve pains were proliferating (5).
But this time, the scientific community had a suspect.
A familiar suspect we have already encountered in many virus investigations.
3. Your immune system is fighting against you
Cytokine Release Syndrome (CRS) had just made its appearance at the top of the list of suspects.
Knowing where to look, scientists started to search for some clues. Tracking elevated concentrations of inflammatory cytokines and chemokines, they found that IL-6, IL-2, IL-1β, IL-8, IL-17, IFN-γ, TNF-α, IP10, MCP-1, IL-10 and IL-4 were all present in COVID-19 patients. There was no doubt, CRS was involved (6).
This disproportionate immune response from the host becomes unfavourable, deleterious, and leads to the failure of several vital organs and death.
But something even more surprising was recently revealed at the end of April.
During any viral infection, our immune response produces the first type of interferon - IFNa. This cytokine has the potential to protect patients and has consequently been used to treat hepatitis B and C.
In the particular case of COVID-19, scientists found that interferon alpha may induce ACE2 gene expression. Since ACE2 is the receptor of the virus on cells, it would mean that your immune system creates new entries for the virus to get into cells. This discovery is still awaiting confirmation.
More your immune system responds to the threat, more it helps the virus to destroy you.
If lungs were the first organs targeted by COVID-19, due to the high expression of ACE2 on the surface of epithelial cells, it appears that the viral infection triggers Cytokine Release Syndrome.
This immune reaction overdrive leads inexorably to the fall, one after the other, of all the patients’ organs.
Worse than that and due to the overexpression of the ACE2 gene, the cytokine IFNa seems to offer more cell portals to the virus, so accelerating the process of a highly probable death.
The best trick of the virus is being able to turn our proper immune system against us.
Diaclone offers a specific range of Antibodies, ELISA and ELISpot kits for COVID-19 research that can be used to monitor cytokine levels in samples.
Are you working on infectious diseases and want to monitor humoral immune response?
Better try DIACLONE’s products to succeed!
REFERENCES
(1) Retrospective study on the first 191 patients in Wuhan, China. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study – Fei Zhou, Ting Yu, Ronghui Du.
(2) A Novel Coronavirus from Patients with Pneumonia in China, 2019 - Na Zhu, Ph.D., Dingyu Zhang, M.D., Wenling Wang, Ph.D., Xingwang Li, M.D., Bo Yang, M.S., Jingdong Song, Ph.D., Xiang Zhao, Ph.D., Baoying Huang, Ph.D., Weifeng Shi, Ph.D., Roujian Lu, M.D., Peihua Niu, Ph.D., Faxian Zhan, Ph.D., et al.
(3) Human Kidney is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection – Bo Diao, Chenhui Wang, Rongshuai Wang, Zeqing Feng, Yingjun Tan, Huiming Wang, Changsong Wang, Liang Liu
(4) Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China - Dawei Wang, MD; Bo Hu, MD; Chang Hu, MD; et al
(5) Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China- Ling Mao; Huijuan Jin; Mengdie Wang; et al
(6) Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? - Bingwen Liu, Min Li, Zhiguang Zhou, Xuan Guan, and Yufei Xiang
(7) SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues - Carly G.K. Ziegler, Samuel J. Allon, Sarah K. Nyquist, Alex K. Shalek, Jose Ordovas-Montanes
Covid-19 Latest News
The obstacle course of serological tests, viral reinfection, and reactivation.
With more than 3,000,000 cases and 200,000 deaths, and after three months of worldwide outbreak, scientists keep on learning about COVID-19 every day.
How can people be positively diagnosed twice to COVID? Are serological diagnostic tests 100% reliable? Which part of the population is more likely to be infected?
Let’s review the latest updates on what we know about this challenging virus, and the on-going assay developments.
1. The obstacle course of serological tests
The last few months have seen the fast emergence of rapid molecular tests to test presence of virus RNA in patients. Today, it’s the turn of serological tests to take centre stage. With serological tests, the aim is now to measure host immunity to the virus. But the game isn't won yet, and many challenges stand in the way of success:
- Specificity: Many seasonal coronavirus (HCoV-OC43, HCoV-HKU1, HCoV-229E, and HCoV-NL63) that cause colds circulate around the globe. They are not genetically identical to COVID-19, but sufficiently enough to give a false positive in case of low specificity tests.
- Sensitivity: A test with low sensitivity - despite a great specificity for the COVID-19 strain - could miss a weak immune response, and so provides a false negative result. A Chinese cohort of 175 patients demonstrated that immune response was highly variable and undetectable in 6% of the cases (Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications - Fan Wu, Aojie Wang, Mei Liu, Qimin Wang, Jun Chen).
- Timing: If people have been infected less than 12 days ago, the seroconversion mechanism hasn’t occurred yet. You’re just measuring no biological material at all during the incubation phase.
- Individual targeted epitopes: It’s also possible that some individuals are developing antibodies against a different viral protein than the one of the test. Nevertheless, it has been reported that antibodies targets are mainly full-length S protein and its receptor-binding domain (A serological assay to detect SARS-CoV-2 seroconversion in humans, Fatima Amanat, Daniel Stadlbauer, Shirin Strohmeier).
- And finally: you need to show the relationship between seroconversion and seroneutralization. If patients are producing specific antibodies against COVID-19, in sufficient amounts, against the tested epitope, how can we be sure these antibodies are protective?
2. All men are created equal. Are you sure?
- Hypertension, cardiovascular diseases, diabetes mellitus, chronic obstructive pulmonary disease (COPD), and chronic kidney disease are aggravating factors of COVID-infected patients’ conditions (Emami A, Javanmardi F, Pirbonyeh N, Akbari A. Prevalence of Underlying Diseases in Hospitalized Patients with COVID-19: a Systematic Review and Meta-Analysis. Arch Acad Emerg Med. 2020;8(1):e35).
- Men are more sensitive than women (Based upon the data of the WHO website, 63% of all deaths were men). Oestrogen concentration? X chromosome? For the moment, we don’t have a clue about the root cause of such a difference, but this question should be addressed in the context of vaccine development.
- Children seem to be less affected by COVID-19 than adults. In a large report gathering 72 314 cases in China, only 2% were under 19 years old. Another report confirms that no ICU admissions or deaths were reported among persons aged ≤19 years in the USA (Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China. Severe Outcomes Among Patients with Coronavirus Disease 2019 (COVID-19) - United States, February 12-March 16, 2020). How can we explain this fact? One hypothesis is that children present less ACE-2 receptors and so less potential cell entries for the virus. We may also suggest that a young immune system doesn’t present the same features as a mature one. The level of antibodies could be lower and the risk of cytokine storm syndrome decreased.
3. Positively diagnosed to COVID-19. Not once, but twice!
Recently, South Korea reported some cases of cured patients tested positive to Covid-19. The Korea Centers for Disease Control and Prevention explained that of 7,829 people who have recovered from coronavirus, 2.1% were positively retested (163 cases).
How is this possible?
- One hypothesis is reactivation. The coronavirus could remain latent in the body and attack the respiratory organs again once it is reactivated. So far, this hypothesis has not been proven, and experts of HIV and HPV are not really in favour of such an option.
- Another explanation lies in the accuracy of diagnostic kits:
- Poor quality of the components of the test can be blamed. Nevertheless, regarding the offer on the market for such tests, and the level of expertise in South Korea in biotechnology field, we can be confident that the country has access to a large number of high-quality grade tests.
- Mutation of the virus may be an option. In fact, it’s unlikely that COVID-19 has already mutated in a drastic way. What’s in it for COVID at the moment if it mutates? It is spreading so easily among the worldwide population, there’s no necessity to change what already works pretty well.
- We might also suggest that fragments of the virus remain in the body. In fact, despite the seroconversion and the cured state of the patient, some residual virus RNA can persist in the bloodstream.
- Reinfection is another possibility. Some people with underlying weak immune system conditions, and presenting lower antibody levels, could face a second infection to the virus. Nevertheless, regarding the antibody immune response of patients, the probability of such event is estimated as being low.
It is clear that we are still in the early stages of our coronavirus discoveries. The obstacles of accuracy in serological tests are numerous, some subset populations present higher risks and the viral reactivation is not so clear for the moment.
However, we need to make decisions without full knowledge of the immunity of COVID-19. The stakes are too high to sit back and do nothing…
To effectively monitor Covid-19 infection & cytokine storm syndrome, DIACLONE has developed highly specific ELISA, Multiplex and ELISpot kits, already validated in a large number of studies.
Fast track your COVID-19 research with Diaclone!
The therapeutic potential of anti-CD25 antibodies
CD? Did you say CD?
In immunology, it is really useful to identify the different types of leukocytes within a sample. But distinguishing leukocytes according to their morphological or functional characteristics is not an easy job.
However, scientists discovered that leukocytes express on their surface a large number of molecules which can be used as markers for their specific identification. The concept of cell immunophenotypic identification was born.
This identification is based on the detection of these membrane markers using monoclonal antibodies followed by analysis in flow cytometry. Since 1982, these markers have been designated according to the cluster of differentiation nomenclature or CD, followed by a number. And the combination of present (CD+) and absent (CD-) markers allows the identification of the state of maturation of the cells.
CD25: a difficult youth
As early as February 1988, a Diaclone monoclonal antibody anti-CD25, the B-B10 clone (now known as Inolimomab or Leukotac) was used in vivo to treat a severe case of acute Graft versus Host disease.
CD25, also known as the interleukin-2 high-affinity receptor alpha chain (IL-2Rα), has been observed on activated B, T lymphocytes and macrophages. Alternative names for the antigen are often used: IDDM10, IL2R, TCGFR, p55 or Tac.
Inhibiting the binding of IL-2 to CD25, anti-CD25 monoclonal antibodies were first developed as therapeutic agents to treat organ transplant rejection (Basilixumab, a Novartis chimeric mouse-human CD25 antibody, 1998 approval) and multiple sclerosis (Daclizumab, a Biogen humanized CD25 antibody, 1997 approval then withdrawn due to safety issues).
But since - as CD25 is constitutively and highly expressed on Treg cells - this marker was rapidly investigated as a potential target for inflammatory, auto-immune and cancer immunotherapies.
In 2013, ADC Therapeutics and Genmab partnered to develop a new antibody format, an Antibody Drug Conjugate (ADC), offering anti-CD25 a second life, this time in the oncology field. Camidanlumab was in the pipeline.
But tackling cancer with a Treg cell depletion tactic - like companies such as Hifibio Therapeutics have promoted since the beginning - was quite a contested therapy approach. So far, clinical trials have proven these strategies wrong. Until recently…
CD25: an oncology boat with the wind in its sails
- Anne Goubier and her team at Tusk Therapeutics made the headlines when they showed that a blockade of IL-2 signalling limits Teff responses. They consequently decided to develop a new version of anti-CD25 antibody that does not block the IL-2 signalling in order to deplete Treg and stimulate Teff at the same time. This promising antibody is going to be tested in cancer. But not by Tusk itself. At the end of 2018, Roche paid €70 million ($81 million) upfront to buy Tusk Therapeutics and in doing so getting complete control on this promising anti-CD25 antibody.
- In 2019, NIH conjugated a near infrared silica-phthalocyanine dye to a monoclonal antibody. This process led to a new cell-specific cancer therapy that locally kills specific cells in the tumour. In this particular context, comparing full length anti-CD25 antibody with its fragment version, it appears that the absence of the Fc portion leads to faster clearance and therefore promotes a superior activated T cell response in tumours.
- January 2020, Alderaan Biotechnology, a preclinical company focused on the development of anti-CD25 monoclonal antibodies for the treatment of cancer, announced that it raised €18.5 million in a Series A financing.
And any recent updates on dysregulated inflammatory response?
In 2019, a Japanese team from Kyoto University and Asahi Kasei Corporation paved the way to a new approach. Apheresis is a technique that makes it possible to take, via a machine, one or more blood components depending on the needs. The team demonstrated that the removal of Tregs from septic patients by apheresis was a good relief solution. However, the removal of cells other than Tregs caused the adverse effect of lowering the immune response against microbes. So, they decided to immobilise an anti-CD25 antibody on a polyethylene fibre to specifically remove Tregs without impacting the other components of PBMCs.
We might agree that it is not a definitive therapeutic solution like those described above for cancer, but this nice creativity - combining immunoaffinity purification with the apheresis technique - must be applauded and encouraged!
Do you have a current project on CD25? Do you want to specifically target Treg?
Diaclone has developed several anti-CD25 monoclonal antibodies for your research:
Clone B-B10: anti-IL2Ra, a biologically active mAb
Clone B-F2: anti-IL2Ra
Clone B-G3: anti-IL2Ra
CD25 ELISA Pair:
Clone B-G3: Capture antibody
Clone B-F2: Revelation antibody
The Dark Side of CAR-T Therapy: CRS
The most common and potentially severe toxicity seen across trials using immunotherapies such as monoclonal antibodies, bispecific antibodies (Bi-specific T-cell engagers or BiTEs) and adoptive T-cell therapies (e.g. CAR-modified T-cell) is CRS.
But what does CRS mean?
CRS stands for Cytokine Release Syndrome. Cytokine Release Syndrome is a clinical syndrome resulting from generalized immune activation correlating with marked elevations of serum inflammatory markers and cytokines. The first clinical signs of CRS are fevers, myalgias, and fatigue. But starting from fever with or without constitutional symptoms (CRS grade #1), symptoms can rapidly evolve to hypoxia requiring O2 (grade #3) and finally death (grade #5).
But is the probability of a CRS event really alarming?
The risk of CRS is influenced by factors related to the type of therapy, underlying disease, and characteristics of the patient. Treatment with most conventional monoclonal antibodies carries a relatively low risk of CRS, whereas CAR-T therapies carry a significantly higher risk of CRS incidence.
With regard to recently approved CAR-T therapies, Yescarta was studied in 107 adults with large B cell lymphoma and ninety-four percent of treated patients developed CRS and 12% developed grade ≥3 CRS. Kymriah was tested in 68 paediatric and young adult patients with relapsed/refractory ALL and seventy-nine percent developed CRS with 49% developing grade ≥3 CRS!
But what can we do when CRS occurs?
Corticosteroids may be necessary in some cases, but in high doses have proven to have a detrimental effect on CARTs.
Extracorporeal blood purification techniques, such as high-volume hemofiltration, cascade hemofiltration, plasma exchange, and coupled plasma filtration adsorption, have also been reported as a potential alternative to treat CRS syndrome. But they remain time consuming and labour extensive solutions.
Since the initial observation that tocilizumab, an antibody against the IL-6 receptor, rapidly reversed severe CRS, the drug remains at the forefront of CRS treatment.
For the future, many CARTs in development incorporate suicide targets as methods to mitigate toxicity. Another approach is manufacturing CARTs that can be regulated without actually killing them, for example requiring infusion of an additional agent for activation.
But diagnosis is better than treatment.
Early prediction is necessary as the side effects appear rapidly - within 1 to 2 weeks - following CAR-T infusion. The elevation of the signature set of 24 different cytokines is then an interesting and crucial measure to monitor in patients.
The main cytokines implicated in the pathogenesis of CRS include effector cytokines released from activated T cells, but also cytokines mainly secreted by monocytes and macrophages. IFN-γ and sgp130 levels, for example, rise early and their elevation is different for severe versus non-severe CRS.
But we can also mention sIL6R, interleukin-6 (IL-6), interleukin-10 (IL-10), interferon INF-α, chemokines that are chemotactic for monocytes/macrophages (MCP1, MIP1α, MIP1β), granulocyte-macrophage colony-stimulating factor (GM-CSF), but also TNF-α, IL-1b, IL1RA, IL-2, sIL2Rα, C-reactive protein (CRP) and IL-8. Other biomarkers of endothelial cell activation, such as Angiopoietin-2 and von Willebrand factor, have also been recently described to predict CRS severity.
In order to effectively monitor the risk linked to immunotherapy, Diaclone has developed a large panel of standard recombinant cytokines and monoclonal antibodies targeting specific cytokines engaged in Cytokine Release Syndrome, as well as a large range of ELISA kits.
Biologically active antibodies
Question: What am I? Antagonist, inductive, repressive, blocking or neutralizing.... Answer: Biologically active antibodies!
They are sometimes qualified as antagonist, sometimes as agonist. Sometimes named inductive, sometimes repressive, sometimes blocking, or sometimes neutralizing. But they are always called antibodies. Have you ever guessed what it was about? Yes, that’s them, the biologically active antibodies!
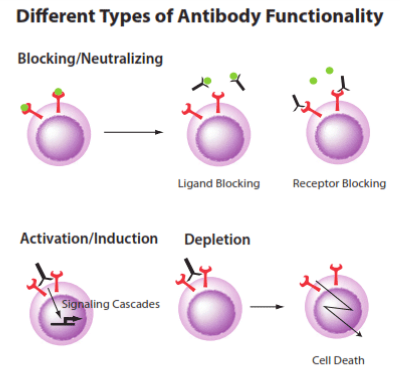
Depending on the nature of the antigen, a specific antibody can demonstrate different biological functions:
- if the target is a cell surface marker for instance, the desired effect of the mAb may involve proliferation (induction/activation effect through a signaling cascade ), inhibition, cell maturation or even the killing of the target cell. Cell depletion will then occur through the recruitment of immune mediators with the Fc portion of the mAb to trigger antibody-dependent cellular cytotoxicity (ADCC), antibody-mediated phagocytosis cytotoxicity (ADCP) or complement-dependent cytotoxicity (CDC).
- if the target is a soluble molecule such as plasma protein (TNF, VEGF…) or a drug, the binding may trigger a blocking effect. When bound to the mAb, these drugs are not able to interact with their normal targets anymore. Of course, blocking effect can also occur via the cell surface receptor like the well-known immune checkpoint inhibitors (targeting CTLA-4, LAG3, PD1 and PDL1).
- If the target is an infectious organism, the desired function of antibodies may be neutralization of the foreign host, so disabling the virus, bacteria or other. In fact, most licensed vaccines teach the body how to make neutralizing antibodies.
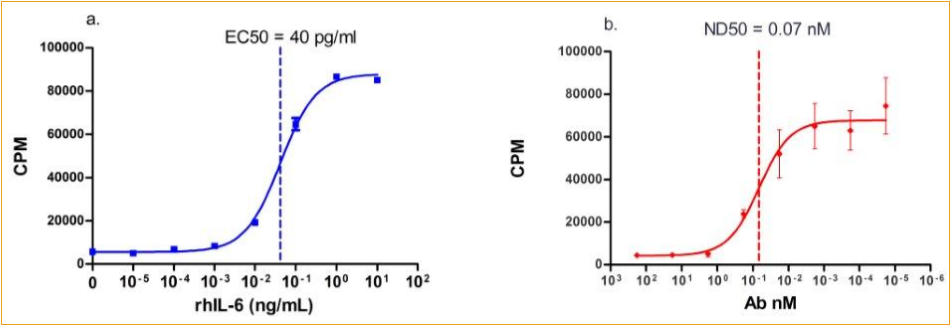
Cell proliferation induced by IL-6 and Neutralization by anti-IL-6 antibody
(a) Recombinant human IL-6 stimulates proliferation in XG1 cell line in a dose-dependent manner. The ED50 is typically 30-40 pg/mL.
(b) Proliferation elicited by rhIL-6 (1ng/ml) is neutralized by increasing concentrations of human IL-6 monoclonal antibody. The ND50 is 0.07nM .
Unfortunately, many difficulties can appear during the process of raising biologically functional antibodies. We can name:
- epitope masking by variable loops or multimerization event,
- the presence of cryptic binding domains,
- the very high binding affinity of the target for its receptor, which makes the generation of inhibitory antibodies extremely challenging,
- the very narrow window for neutralizing antibodies to act before the establishment of a virus infection and its genome integration into the cell host.
Either from a classical hybridoma approach or by phage display technique, DIACLONE has over the years learned to tackle all these obstacles to offer you the best possible antibody references. Thanks to its bioassay platform, all our references are highly validated via proliferation, cytotoxicity, neutralization or various cell functionality tests.
Want to know more about these unique antibody references?
Or you desire to validate your antibodies via our bioassays?
Don’t hesitate to contact us !
What is the KD of my antibody?

KD is the equilibrium dissociation constant, a calculated ratio of koff/kon, between the antibody and its antigen. The association constant (kon) is used to characterize how quickly the antibody binds to its target. The dissociation constant (koff) is used to measure how quickly an antibody dissociates from its target.
KD and affinity are inversely related. A high affinity interaction is characterized by a low KD, a fast recognizing (high Kon) and a strong stability of formed complexes (low Koff).
.png) 10-4 to 10-6 Micromolar (μM)
10-4 to 10-6 Micromolar (μM)
10-7 to 10-9 Nanomolar (nM)
10-10 to 10-12 Picomolar (pM)
10-13 to 10-15 Femtomolar (fM)

At Diaclone, we measure using the SPR (Surface Plasmon Resonance) technology and the Octet instrument. This technique can help you to rank your antibodies but also to better understand the performance of your antibodies.
For example, this ELISA development showed that the pair worked only when mAb1 is coated (Fig.1)
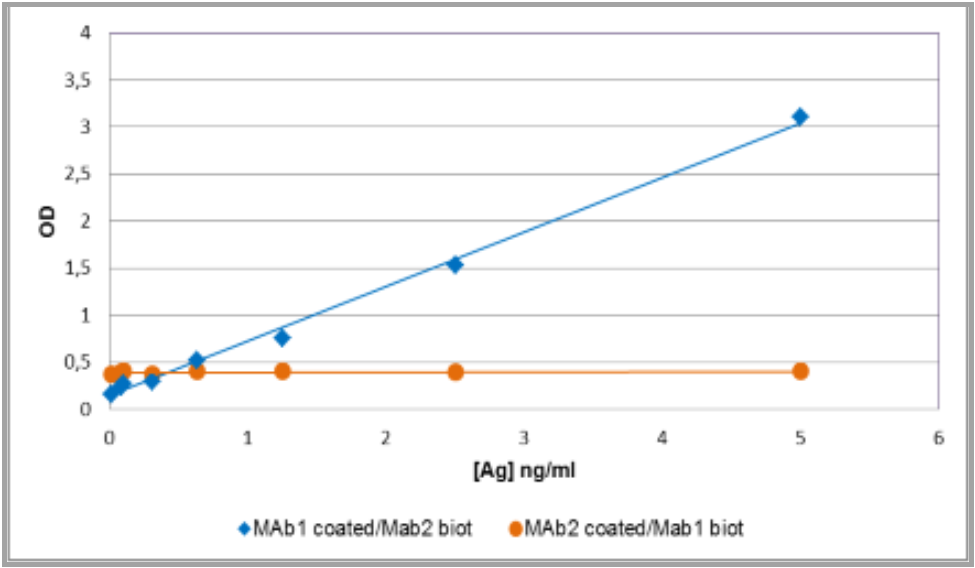
Fig. 1 : mAb1-mAb2 pairing evaluation
The determination of KD and association/dissociation profiles of antibodies by Octet permitted us to understand why the pair works only in one way.
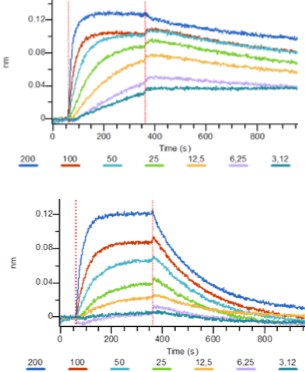
Fig. 2 : Profiles of association dissociation of mAb1 (top) and mAb2 (bottom)
The mAb2 has a quick dissociation time (Fig. 2), so, when it’s coated and after all the washing steps, the antigen doesn’t stay on mAb2. Using this antibody as revelation avoids this dissociation which is linked to accumulated washing steps.
Octet analysis can also help to validate the accessibility of tags (HIS, GST…), to study the interaction receptor-ligand and the potential inhibition of interaction with an antibody, and to perform epitope binning (also known as epitope mapping or pairing).
The advantage of Octet compared to the highly popular Biacore technique is the lower price, the rapid execution of experiments and the small amount of proteins required.
Whatever the interaction to study, Octet is a valuable tool.
Use Diaclone’s expertise to further characterize your antibody
The Octet technology is fully integrated into our custom monoclonal antibody development so that it can be implemented to further study monoclonal antibody candidates.
A large range of custom services, dedicated to your needs:
· Biological activity: agonist or antagonist, secretion enhancer, blocking signal transduction, cellular growth activation or inhibition,….or any new model to design
· Effector activity: ADCC, ADCP or CDC
· Antibody applications: ELISA, Western Blotting, Flow cytometry…
· Antibody labeling
Contact us to discover how Diaclone can support your activity

Happy 30th Birthday Diaclone ELISpot!

A little bit of history ...
In 1983, two laboratories on opposite sides of the world described almost simultaneously what would later be called the ELISpot: Cecil Cervinsky in Gothenburg, Sweden and Jonathon D. Sedgwick in Perth, Australia. This “invention” was first dedicated to enumerate frequency of B hybridroma cells secreting an antigen specific immunoglobulin and then, a few years later, to measure the frequency of T lymphocytes secreting a specific lymphokine.
At that time, Monoclonal Antibodies were rare, the ELISA was still a very new technique and the specificity of most polyclonal antibodies was highly questionable, but with the proximity of the team of Dr Outcherlony at Gothenburg University, Cecil Cervinsky had the idea to imagine an “Antibody forming cell ELISA assay” in a gel matrix.
Since the first description, key advances have included the first use of membrane bottomed plate (nitrocellulose and then PVDF) that enables increased sensitivity, improvement in substrates and application of computer-assisted spot counting technologies (1993) and the commercialization of these technologies by numerous instrumentation companies.
The assay was called by a range of names including spot-ELISA, ELISA-spot, ELISA-immunospot and, of course, ELISpot ultimately chosen by the scientific community.
Diaclone’s ELISpot ...
ELISpot is now the method of choice in clinic for monitoring Interferon-gamma secretion in cancer immunotherapy and viral vaccine trials.
The Diaclone IFNg ELISpot assay is used in several clinical studies and published in Clinical Cancer Research by Dosset et al.,Godet et al. and Teixeira, L. et al.*
Diaclone ELISpot Kits and ELISpot Sets
Diaclone ELISpots (Enzyme-Linked Immunospot Assays) are highly specific immunoassays for the analysis of cytokine and other soluble molecule production and secretion from T-cells at a single cell level in conditions closely comparable to the in-vivo environment with minimal cell manipulation.
Utilising sandwich immuno-enzyme technology, Diaclone ELISpot and Dual ELISpot assays can detect both secreted cytokines and measure the frequency of single cells that simultaneously produce multiple cytokines or other effector molecules.
Formats available
- ELISpot Kits: Pre-coated PVDF plate(s), Detection antibody, Alkaline Phosphatase conjugate, BSA and BCIP/NBT ready- to-use substrate buffer.
- ELISpot Sets: Capture and Detection antibody, Alkaline Phosphatase conjugate, BSA and BCIP/NBT ready-to-use substrate buffer.
- ELISpot Pair: Capture and Detection Antibodies.
- Dual ELISpot Sets: Capture and Detection antibodies for two analytes, Alkaline phosphatase conjugate, blocking reagent, Ready-to-use BCIP/NBT substrate buffer, Peroxydase conjugate, BSA, Ready-to-use AEC substrate buffer.
- Dual Fluorospot Sets: Capture antibody for two analytes, FITC-conjugated Detection antibody for cytokine 1, anti-FITC antibody green fluorescence conjugate, biotinylated detection antibody for cytokine 2, streptavidin-phycoerythrin conjugate, BSA.
Valuable Investigative Tool
ELISpot techniques are amongst the most-sensitive methods available (up to 400x more sensitive than conventional ELISA) for cytokine research and benefit from a technically easy performance, rapid detection time and no requirement for expensive equipment or analysis software.
High Performance
 Highly Sensitive assay can allow the detection of a single cell out of 100,000
Highly Sensitive assay can allow the detection of a single cell out of 100,000- Fast procedure following cell stimulation
- Accuracy and reliability are guaranteed as all our reagents have been validated according the ISO 9001:2000 quality systems
- Reagents generate well focused, defined and easy to analyse ‘spots’
- No cross reactivity with other human cytokines tested
Flexibility
- Extensive kit range across different species with high specificity and affinity for a number of different cytokines and soluble molecules
- Mono and Dual cytokine analysis available using both enzymatic and fluorescent detection systems
- ELISpot reagents available in a number of formats from whole pre-coated kits for easy analysis and increased throughput to matched Antibody Pairs a great tool for developing research
The CAM Family
A NEW target for monitoring or treating cancers?
The cell adhesion molecules (CAMs) family includes more than 50 proteins with four main groups: immunoglobulin (Ig)-like CAMs, cadherins, selectins, and integrins.
Many cellular functions are directly linked to cell adhesion such as signal transduction, cellular communication and recognition, embryogenesis, inflammatory and immune responses, apoptosis and some of them also act as viral receptors (Cohen MB, Am J Clin Pathol. 1997, 107(1):56-63).
The metastatic dissemination of tumor cells is the leading cause of morbidity and mortality in patients with cancer since it designates the transition from a localized, potentially curable to a generalized, usually incurable disease (Makrilia N, Cancer Invest. 2009, 27(10)).
Across the years, it has become evident that the adhesion properties of neoplastic cells play a pivotal role in the development and progression of cancer. (Okegawa T, Acta Biochim Pol. 2004;51(2):445-57)
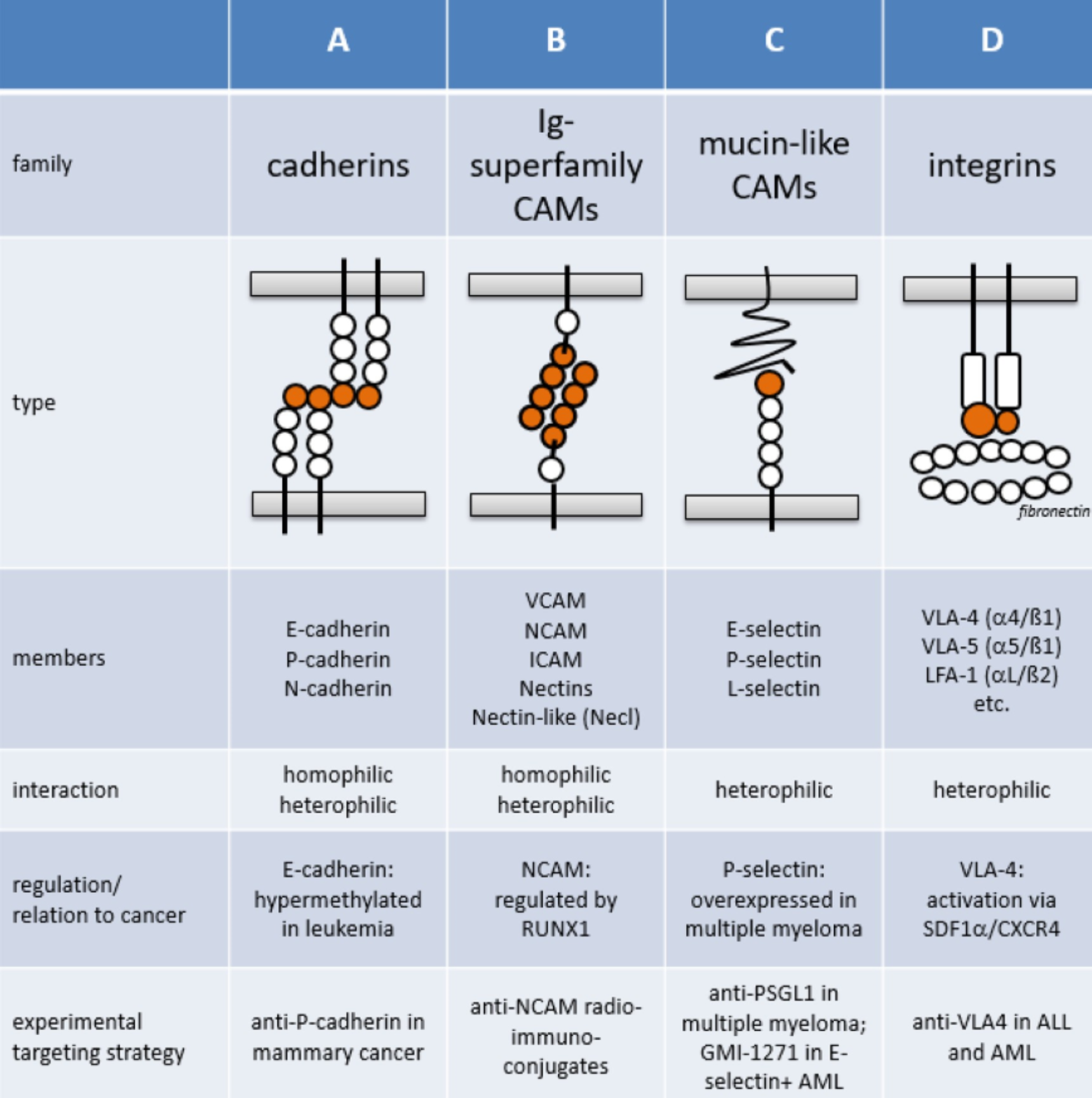
(Windisch R, Cancers 2019, 11(3), 311)
Changes in the expression or function of CAMs have been associated with alterations in the adhesive or signalling status of tumor cells, allowing them to acquire a more motile and invasive phenotype prognostic biomarkers or as potential therapeutic targets in malignancies.
Additionally, many of CAMs can be cleaved and released by proteolytic cleavage activity, and their soluble forms were found to be increased in serum levels of cancer patients. Even if elevated levels of soluble CAM are also observed in bacterial and viral infections or in acute inflammation, some of them have been identified to be interesting prognostic markers of cancer progression, such as EpCAM, described to be upregulated in colorectal cancer with clinical relevance (Han S, Ebiomedicine 2017; 20:61–69).
Diaclone has been interested for many years in the adhesion molecules and can provide antibodies against all of the selectin and integrin families, most of IgSF CAM family and against EpCAM, H-CAM, M-CAM, and BL-CAM.
Knowing that the soluble form levels could become an innovative tool of cancer monitoring, Diaclone has also developed ELISA kits for measuring serum levels of a wide range of sCAMs.
How can Diaclone support your CAR-T development? |
|
|
Diaclone continues its implication in innovative health technology with its participation in the development of IL-1RAP CAR T cells (Warda W Cancer Res February 1 2019 79 (3) 663-675) and in a CD123 CAR T cells project (both in collaboration with the INSERM U1098 research team in Besançon, France). Diaclone is renowned as a specialist in antibody development - in particular CD markers and membrane expressed proteins. Diaclone’s competence in cell biology and antibody development coupled with its new molecular biology platform and the antibody engineering capabilities of the mAbexperts team (www.mabexperts.com) can provide:
|
|
|
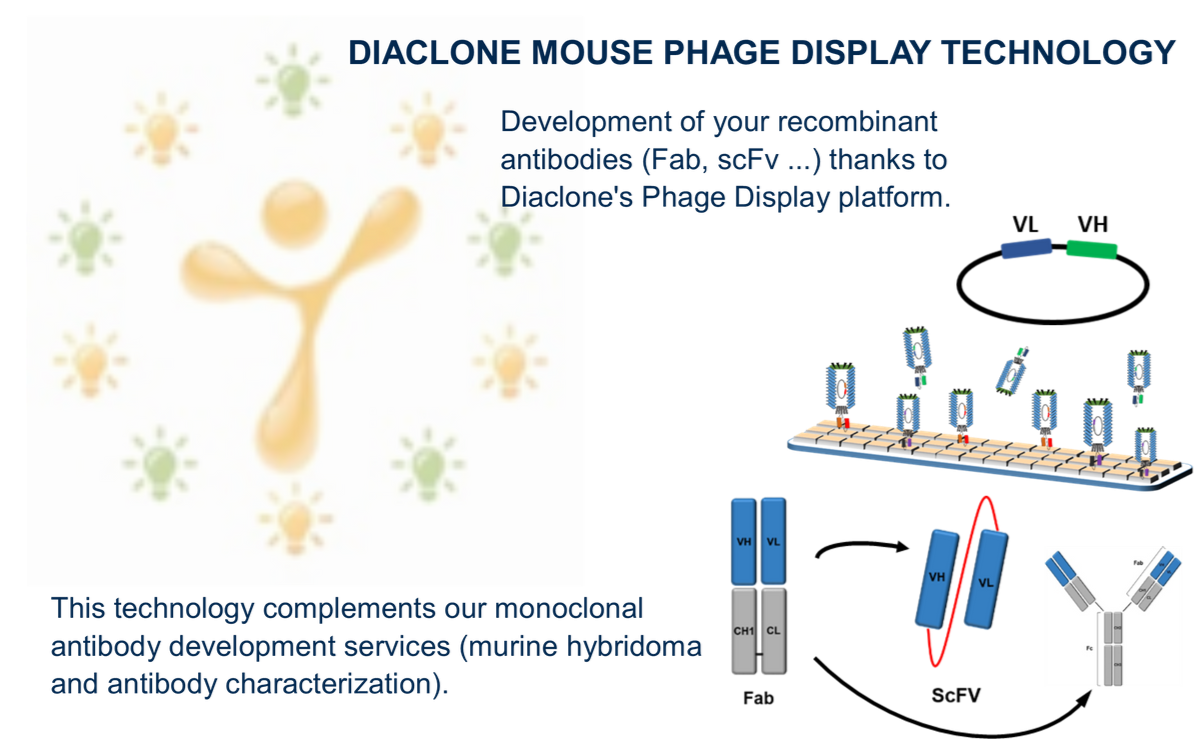
Diaclone mouse phage display platform, an alternative approach to mAb development
A case study:
Diaclone, with over 30 years of experience and expertise in immunology products, offers a vast catalogue of mAbs and ELISA kits for use in research and diagnostic applications. A large number of Diaclone’s products are cytokine related and specifically Interleukins (IL).
The development of the anti-IL-8 antibody was initiated in 1991, according to Köhler and Milstein’s technique (Nature-1975, 256, p495-7); six fusions were carried out giving only one specific anti-IL-8 mAb, the clone B-K8, which is still used in Diaclone’s IL-8 ELISA kit paired with a rabbit polyclonal. The aim of this project was to replace the polyclonal.
At the beginning of 2019, a new project of anti-IL-8 mAb development was initiated with a new IL-8 mouse immunization. One part of the cells from the immunized animals was unsuccessfully fused using the classical mAb method and the second was processed by the Diaclone Phage Display Platform. The phage display method (Smith, Science-1985, vol. 228, n°4705, p1315-17) is an alternative way of developing mAbs using molecular biology. The platform is capable of obtaining a strong Fab library of IgG sequences from spleen cells coming from immunized mice.
The library obtained was screened in biopanning steps with IL-8 to select very specific clones. One hundred selected Fabs were tested in ELISA to keep the best candidates. Ten of them were selected and evaluated in pairs with B-K8. Finally, we obtained four good substitutes to the polyclonal anti-IL-8. These four Fabs were sequenced and reformatted by molecular biology in full IgG to be further validated in the IL-8 Elisa kit. As a result of this project, phage display technology has demonstrated its interest, notably in allowing mAb development which was particularly difficult to obtain with a classical fusion.
Case Study Poster available here
Assays for the Human IL-17 Family
It has been demonstrated that T helper cells can differentiate into IL-17 producing cells independent of Th1 or Th2 pathways establishing Th17 cells as a unique T-cell lineage challenging the classical two pathway theory.
This new T helper cell lineage is defined by its ability to produce IL-17A, IL-17F and IL-17A/F.
Diaclone has developed a large range of ELISA and ELISpot kits for the detection of Human IL-17A, IL-17F and IL-17A/F.
Read more about this product range